Abstract
Despite significant advances in cancer microbiome research, advances in identified confounders and absolute quantification of the microbiome remain underutilized. This raises concerns about the possibility of false associations. Here we examine the fecal microbiota of 589 patients at various stages of colorectal cancer (CRC) and compare our observations with up to 15 published studies (4,439 total patients and controls). did. Using quantitative microbiome profiling based on 16S ribosomal RNA amplicon sequencing combined with strict confounder control, we determined that transit time, fecal calprotectin (intestinal inflammation), and We identified BMI as the primary microbial covariate.
Well-established microbiome CRC targets such as Fusobacterium nucleatum were not significantly associated with CRC diagnostic group (healthy, adenoma, carcinoma) when controlling for these covariates. In contrast, the relevance of Anaerococcus vaginalis, Dialister pneumosynthes, Parvimonas micula, Peptostreptococcus anaerobius, Porphyromonas asacharolytica and Prevotella intermedia remains robust and could be considered for future targets. It emphasizes the possibility of Finally, control subjects (age 22–80 years, mean 57.7 years, standard deviation 11.3) who met the criteria for colonoscopy (e.g., positive fecal immunochemistry test) but did not have colonic lesions; The intestinal dysbiosis is assigned to the Bacteroides 2-enriched enterotype. Highlights the uncertainty in defining healthy controls in cancer microbiome research. Taken together, these results highlight the importance of quantitative microbiome profiling and covariate control for biomarker identification in CRC microbiome studies.
Mainly
The incidence of colorectal cancer (CRC) is steadily increasing1, especially in people under 50 years old2. It is estimated that CRC3 causes approximately 16 deaths per 100,000 people in the United States and 14 in Belgium each year. Identification of individuals at high risk is essential, as medical interventions can effectively reduce colorectal cancer progression and associated mortality.
Colonoscopy with adenoma polypectomy reduces the risk of colon cancer by up to 90%4. Early identification of individuals with polyps would reduce the global burden of colorectal cancer. Nevertheless, identifying patients at increased risk remains a challenge, highlighting the need for population-wide screening.
Changes in the microbiota are associated with various disease phenomena5. Several bacterial markers, such as Fusobacterium , have been found to be abundant in the lesions and stool of CRC patients in developing and developed countries, 15 suggesting a possible microbiome-based role. . Diagnosis and/or prognosis suggested.
Microbiome profiles are influenced by multiple variables that can perturb or amplify biological phenomena, but covariate control is far from the norm. For example, water content, an indicator of transit time, remains uncontrolled despite having the greatest explanatory power for overall gut microbiota variation in some cohorts 16,17. Intestinal inflammation is measured as fecal calprotectin, which reflects increased neutrophil excretion into the intestinal lumen, and is more sensitive than fecal occult blood in identifying CRC patients, 21 and molecular fecal CRC screening. 19 has become a potentially untapped target.
Relative microbiome profiling (RMP, where taxon abundance is expressed as a percentage) remains the primary approach in microbiome research. However, given the issues regarding composition22 and relative profile interpretation23, the use of experimental and quantitative approaches is increasingly recommended23,24,25. This reduces both false-positive and false-negative rates in downstream analyses, thereby reducing the risk of misinterpretation of microbiome associations and focusing clinical programs on biologically relevant targets. Can be 25. Although quantitative microbiome profiling (QMP) allows normalized comparisons between different samples or conditions, 24,25 no QMP-CRC microbiota studies have been performed to date. In this article, we address these two gaps in CRC microbiota research: (1) quantitatively characterizing the microbiota profiles associated with colonic malignant transformation and (2) the biology behind the microbiota. Identifying covariates in the microbiome that represent clinical phenomena - potentially obscuring CRC associations. To achieve this aim, we examined the microbial profiles of 589 Belgian patients from the University of Siekenhuis Leuven (UZL) who justified colonoscopy based on clinical symptoms, including patients with colorectal cancer. and compared these with existing public datasets (total n = 4,439). patients and controls). To our knowledge, this is the first large-scale study of the gut microbiota at all stages of colorectal cancer development, combining QMP analysis with a comprehensive analysis of microbiota covariates to It separates related signals from confounding signals and specifically identifies related taxa. You have colorectal cancer.
Result
Intestinal inflammation occurs more frequently in patients with colorectal tumors
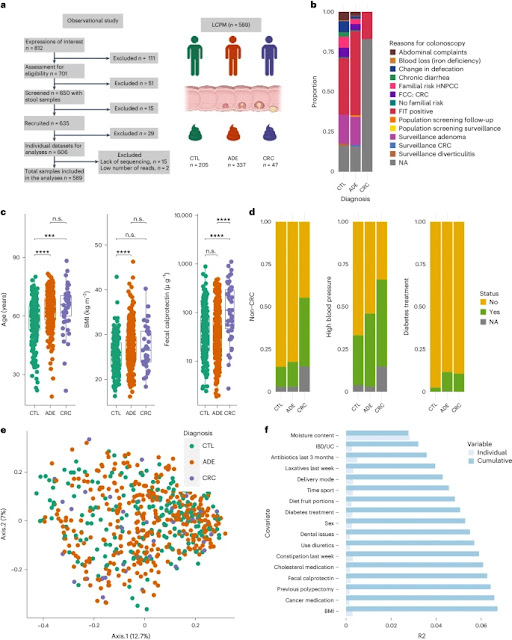
We recruited 650 volunteers from 2017 to 2018 who were referred to UZL for colonoscopy and colectomy and provided a stool sample before the colon procedure. Most of the participants were from the Flanders region of Belgium. In this study, we defined cancer stage as a diagnostic group and divided participants into three groups based on thorough colonoscopy and clinical evaluation: (1) no signs of colonic involvement; patients (CTL, nâ=â205), (2) patients with polyps (n < 10, size 6–10 mm, if polyps are considered precancerous lesions) (ADE, n = 337) and (3) CRC patients (nâ = 47, 2 (4%) stage 0, 14 (30) %) stage I, 13 (28%) stage II, 11 (23%) stage III, 3 (6%) stage IV , 4 (9%) stage undetermined). Patients who did not meet these criteria or had insufficient clinical or molecular data were excluded. The final cohort of the Leuven CRC Progression Microbiome (LCPM) study consisted of 589 patients. The most common indications for colonoscopy were either a positive fecal immunochemical test (FIT) or adenoma surveillance. Other signs include familial risk, abdominal discomfort, and changes in bowel habits (Figure 1a and Supplementary Table 1). This study was registered with his Clinicaltrials.gov (NCT02947607).
We collected a comprehensive set of 165 universal metadata variables (not specific to each of the three groups) from each participant. After curation, variables with collinearity (if Pearson is |r|→0.8, keep variables with less missing data) or variables with incomplete data coverage (more than 20% of variable values are missing) has been excluded. . The final set consisted of 95 high-quality variables (Supplementary Table 2).
To identify metadata variables associated with diagnostic groups, we applied two statistical approaches: (1) the nonparametric Kruskal-Wallis (KW) test for all numerical variables and its Ύ·2 effect size (see Supplementary Table 3), and (2) chi-square (CS) tests for categorical variables and Cramer's V effect size (CV) (Supplementary Table 4) followed by the Benjamini-Hochberg method of multiple testing correction (adjusted P). follows. We analyzed eight variables associated with diagnostic group (false rate <5%): age, body mass index (BMI), calprotectin, reported sleep duration, previous cancer (including colorectal cancer); Discovered the condition of the tooth (complete, partial). ), diabetes treatment and hypertension (Supplementary Tables 3 and 4). CTL patients were younger (n = 589, KW test, β2 = 0.058, 2 = 35.77, adjusted P = 2.6). ×10:7; post-hoc Dunn test (PhD), adjusted P≡0.05 for CTL compared with ADE or CRC groups) and lower BMI (n≠). 553, KW test, Ύ2â=â0.023, Ï2â=â15.73, adjusted Pâ=â10â3; test, CTL vs. ADE (adjusted Pâ<â0.05), sleep better than participants in the other two diagnostic groups. Time reported to be shorter (nâ=557, KW test, η2â=â0.019, Ï2â=â13.41, adjusted Pâ) =–4.6–10–3; pHD test, adjusted for CTL vs. ADE P–0.05; see Supplementary Table 3. Water content, an important microbiota covariate 16, was not significant in all diagnostic groups (nâ=â589, KW test, η2â=â0.001, η2â=â1.32, adj. Pâ=â=7.0Ãâ10â1).
Calprotectin levels were positively correlated with malignant transformation. CRC patients showed greater intestinal inflammation as measured by fecal calprotectin 18,26 (Figure 2). 1a and Supplementary Table 3). Notably, CRC showed higher values (219.42 μ g g g 1, range 2.74–1,114.42, n = 47) compared to ADE (70.24 μg g g 1). , range 1.87 to 487.21, n ~ 337) or CTL (73.25 to gâg 1, range 2.42 to 884.82, n ~ 202) (Fig. 1a, N ~ 583, KW test, μ 2 ~ 0.047, μ 2 ~ 29 .43, Adjusted P–3.0–––10–6; adjusted P–<–0.05 for PhD test, CRC vs. CTL and CRC vs. ADE). We also observed an increase in fecal calprotectin in patients who reported previous cancers (primarily breast and prostate cancer) (Wilcoxon rank sum test (WR), W = 11,067, adjusted P = 4.1) . 10–3), taking anticancer drugs (WR test, W–3,671, adjusted P–<–0.05), heartburn symptoms (WR test, W–11,067, adjusted P–1.0–10–10) , decreased fiber content (WR-test, W=20,964, adjusted P=3.3×102).
Chronic disease history varied by diagnostic group. CRC patients had a higher proportion of previous non-CRC cancers (47.5% vs. 15.0% and 12.1%, CS examination, CV 0.24, Ï2≠31.65, d.f. 2, adjusted P‑=). Ï1.98 × 10 × 2) and hypertension (60.0% vs. 44.3% and 30.5%, CS test, CV 0.17, Ï2 × 16.55, d.f. 2, adjusted P × 1.98 × 10 × 2) (Fig. 1b and Supplementary Table 4 ) ). The CTL group had the lowest diabetes treatment (2.4% vs. 10.3% and 10.6%, CS testing, CV 0.15, Ï2 = 13.79, d.f. 2, adjusted P = 1 .98). Ãâ10â2) (Fig. 1b and Supplementary Table 4), and almost complete dental set (53.3% vs. 35.2% and 32.5%, CS test, CV 0.03, Ï2â=â30.78, d.f. of 10, adjusted Pâ=â1.98Ãâ10â2 ) (Supplementary Table 4).
Known confounders, but not diagnostic group, explain overall variation in the microbiome across stages of colorectal cancer development
The influence of microbiota covariates and the quantitative amplitude of observed microbiota changes have not been well studied in CRC. We combined sequencing data with flow cytometry measurements of fecal microbial load23 to generate QMP data from the study cohort23. We used the established ones to examine QMP variation in relation to the 94 potential covariates listed above (95th being microbial load). Principal coordinate analysis (PCoA; Fig. 1c) of the Bray-Curtis dissimilarity matrix (BCD) at the species level revealed no significant separation between diagnostic groups. Furthermore, no difference in total microbial load was detected between groups (n = 589, KW test, 2 = 0.68, adjusted P = 8.2). â10â1). Distance-based redundancy analysis (dbRDA) revealed 24 microbiota covariates associated with microbial diversity in this cohort (Fig. 1d and Supplementary Table 5). We identified 17 nonredundant covariates that collectively explained 6.7% of the variation in microbiota composition (Supplementary Table 5). Consistent with previous reports16,17, water content showed the highest explanatory value (2.8%) of all covariates (n = 589, stepwise dbRDA, R2 = 2.8%, adjusted P = â). 2→10→3). Intestinal disease/ulcerative colitis (IBD/UC) status, a risk factor for CRC and probably related to microbial gut microbiota and intestinal inflammation 27 , was the second largest covariate . IBD/UC explained 0.4% of the microbiota variation (n = 569, graded dbRDA, R2 = 0.4%, adjusted P = 2 x 10). 3). Other top microbiota covariates included antibiotic and laxative use (Figure 1d). Variation due to mode of delivery (caesarean section or natural delivery) is explained to be 0.3% (nâ533, staged dbRDA, R2â0.3%, adjusted Pâ=â2â10â 3). However, this cohort may be confounded by diet (meal proportion). Vegetables; CS test, Ï2â=â33.09, d.f. 14, Pâ=â2.8ÃÃÃ10Ã3, adjusted Pâ<â<â00.05). Intestinal inflammation (fecal calprotectin) was accounted for at 0.2% (n = 583, graded dbRDA, R2 = 0.2%, adjusted P = 2.6 × 10). 2).In contrast to a previous study in a Flemish population (Flemish Gut Flora Project, FGFP)17, age did not explain the variation in the microbiota (nâ=â589, univariate dbRDA, R2â =â0.2%, adjusted Pâ=â5, 9Ãâ10â2). Surprisingly, cancer diagnosis group (CTL, ADE, and CRC) as a covariate was not associated with microbial variation (n = 589, univariate dbRDA, R2 = 0.2%, adjusted P) . =â0.22; Supplementary Table 5)
The association between Fusobacterium and CRC stage disappears when controlling for confounders or using QMP
Microbiota signals may be taxon-specific and therefore do not reflect changes in the overall community. Although various microbial associations have been reported in CRC studies using RMP6, 7, 8, and 13, we used QMP to identify species whose absolute abundances were related to diagnostic groups. Comparisons were limited to 138 species with prevalence >5% in at least one diagnostic group in the LCPM cohort (Supplementary Table 6). Only eight species showed significantly different abundances (absolute or relative) between the diagnostic groups: Anaerococcus vaginalis (Anaerococcus obesiensis), Alistipes onderdonkii, Diarystar pneumosynthes, Fusobacterium nucleatum, Parvimonas micro, and Pep. Streptococcus anaerobius, Porphyromonas asacharolytica and Prevotella intermedia (KW test, adapted) pa). <â0.05; Figure 2a,b and Supplementary Table 7). Fusobacterium nucleatum was consistently associated with colorectal lesions in different background cohorts, and in the LCPM cohort,13,14 the absolute abundance of Fusobacterium nucleatum was positively correlated with high fecal calprotectin levels (Spearman Rank and Kendall tau correlations, adjusted). Pâ<â0.05;Fig. 2c, Extended Data Fig. 1 and Supplementary Table 8) and cancer progression (diagnosis group) (KW test, β2=0.010, adjusted P= 1.84x10â5, pHD test of CTL adjusted P= 8.80x10â1) vs. ADE, CTL vs. CRC adjusted P‒=‒3.84‒10‑7, ADE vs. CRC adjusted P‒=‒3.84‒10‒7, Figure 2c and Supplementary Table 7). However, after clearing up confusion about calprotectin, neither BMI, water content, and calprotectin alone or in combination nor absolute or relative Fusobacterium nucleatum abundance were associated with diagnosis (general linear variance model analysis (ANOVA), n= 547, P). >â0.05;Extended data diagram 2)
Several established microbial CRC markers are associated with transit time, intestinal inflammation, and BMI, but not with CRC stage
The association between Fusobacterium abundance and fecal calprotectin was previously reported by adding water content, the most important covariate of the microbiome, and BMI, the difference between diagnostic groups. We were prompted to investigate the influence of this confounding factor on CRC-associated genera. To achieve this objective, we analyzed 89 species-level samples from 10 published cohorts 6, 9, 11, 13, 14, 28, 29, 30, and 31 (containing 1,633 samples). We created a list of CRC markers and 67 genus-level CRC markers from 15 cohorts. 7, 8, 9, 11, 12, 13, 14, 15, 28, 29, 30, 31, 32 (equivalent to 4,439 samples). This compiled list of taxa was used as a baseline to test whether the CRC associations of these taxa within the cohort were influenced by the target covariates. To reduce the impact of different statistical treatments, species-level microbial profiles for 9 out of 10 studies were downloaded from the curated resource MetagenomicData33 and analyzed using the statistical component of the pipeline. Spearman correlations between taxon abundance and the three focal covariates showed strong associations between microbial targets and these confounders within species (Extended Data Figure 1). 3a) and genus level (Fig. 3b). Most of these associations were replicated in an independent population cohort (FGFP), suggesting that these associations were strong and not specifically associated with CRC (Extended Data, Figure 3) . Water content, a known major covariate in microbiome studies, 17 is unsurprisingly associated with many taxa examined in both cohorts. Figure 3: BMI, gut inflammation and water index
When compiling CRC-associated taxa from non-QMP studies, we performed analyzes using both RMP and QMP to determine whether the association of confounders influenced the quantitative association of a biomarker or target with a diagnostic group of LCPM. I evaluated whether to give it or not. Of the species previously associated with CRC using QMP and RMP, only 8% (6 of 89) and 10% (9 of 89) were replicated after controlling for confounders. was. Anaerococcus vaginalis, Dialister pneumosynthes, Parvimonas micula, Peptostreptococcus anaerobius, Prevotella intermeia and Porphyromonas asacharolitica were identified by controlled QMP and RMP. Control QMP excluded Fusobacterium nucleatum and Alistipes onderdonkii, suggesting that the previous association of these two species may be incorrect (Fig. 3a). We identified eight species previously associated with CRC (i.e. using QMP and/or RMP), including Fusobacterium nucleatum and Peptostreptococcus anaerobius, which are associated with inflammation (Fig. 3 and Supplementary Tables 8 and 9). This association has so far only been reported for three of the eight taxa mentioned above (Escherichia, Fusobacterium, and Streptococcus)24. Further validation of this association was performed using FGFP (Extended Data Figure 3 and Supplementary Tables 8 and 9). Recognizing that inflammation is a risk factor and not a prerequisite for colorectal cancer progression, we further investigated markers associated with diagnostic groups related to inflammatory status. To this end, we focused on a subset of 340 samples that showed normal calprotectin levels (fecal calprotectin <50 μg μg μg (ref. 34)) regardless of CRC status, suggesting a lack of evidence of local inflammation (112 CTL, 216 CTL ADE and 12 CRC). Evaluating the above 89 CRC markers at the species level confirmed the association of three of the six replicating species (Anaerococcus virginis, Prevotella intermedia, and Porphyromonas asaccharolytica) independent of intestinal inflammation (Supplementary Table 10).
Patients undergoing colonoscopy with or without colorectal cancer have an excess of enterotype Bacteroides 2.
To examine her LCPM cohort in a population context, participants were enterotyped using Dirichlet multinomial mixtures (DMM) on a genus matrix against the background of microbial variation observed in FGFP samples (nâ=â1,04517). Following the previous description of the Flemish population, 23 we identified four community types. “Bacteroides1” (Bact1), “Bacteroides2” (Bact2), “Prevotella” (Prev), and “Ruminococcaceae” (Rum). The distribution of enterotypes differed between LCPM and FGFP (CS test, Ï2 = 34.3, d.f. 3, adjusted P = 1.7–107), but no differences were observed between diagnostic groups within the LCPM cohort. (pairwise CS test, adjusted P→0.1). A pairwise comparison of the prevalence of the dysbiosis Bact2 enterotype in the diagnostic groups of the LCPM cohort revealed that this enterotype was enriched in all CRC diagnostic groups compared to the FGFP population. (test of equal or given proportions, FGFP vs. CTL: β2 α = α15). 09, d.f. 1, adjusted Pâ=â1.1?10â4, 93, d.f. 1, adjusted Pâ=â2.4?10â5, FGFP vs. CRC: Ï2â=â4.34, d.f. 3.4×10×2). Although dysbiosis and the development of colorectal cancer have previously been associated,13,35 the high prevalence of this enterotype in LCPM was unexpected, even in samples from patients without lesions. is. Consistent with previous reports 24 , 25 , this group of Bact2 enterotypes has low cell numbers, low cell abundance, increased calprotectin levels, decreased butyrate producers, and proinflammatory bacteria. showed all the hallmarks of dysbiosis, including an increase in.
 |
| Fig. 4: The Bact2 enterotype is enriched in patients referred for a colonoscopy (with and without colorectal lesions) |
Additional categorical variables appear to be associated with Bact2 enterotypes. These included antibiotic intake (CS test, Ï2 = 30.78, d.f. 3, adjusted P = 2.1 10 2), current treatment with antibiotic inflammatory drugs (CS test, Ï2 = 30.78, d.f. 3, adjusted P=) is included. Ï2.1–10–2), diabetes treatment (CS test, Ï2–=–30.78, d.f. of 3, adjusted P–3.3–10–2), recent diarrhea (last week) (CS test, Ï2–30.78 , d.f. of 3, adjusted Pâ=â10â2, history of gallstones (CS test, Ï2â=â30.78, d.f. of 3, adjusted Pâ=â4.7âÃâ10â2), and recent laxative use (last week) (Ï2â=â30. 78, d.f. of 3, adjusted Pâ =â4.2×10â2) (Supplementary Table 11).
Discussion
The association between the gut microbiota and colorectal cancer is widespread, but it is important to use QMP and extensive metadata collection to identify spurious associations between specific taxa and malignant transformation. This is the first study to systematically investigate microbiome covariates that can obscure or create. At first glance, this study revealed a gut microbial profile that is partially consistent with previous reports on CRC-associated taxa. However, further analysis suggested that many of the previously reported associations were confounded by microbiota covariates, including prominent biomarkers such as Fusobacterium (nucleatum). A total of 17 of the 94 variables explained 6.7% of the observed variation. Of these, water content had the highest explanatory power (2.7%), more than 8 times that of the next covariate (IBD status). The explanatory power of fecal calprotectin was low (0.2%) but significant. Age and especially diagnostic group did not.
Some associations were complex in nature. For example, consistent with previous reports, BMI showed an association with both microbial composition17,25 and cancer progression,36 but in this one cohort, we found no association between changes in BMI and cancer progression. Other factors such as suggested age 37 were not significant. Inflammation is known as a risk factor for CRC38, but its influence on the formation of cancer-associated microbiota remains to be elucidated. Fecal calprotectin is well documented as a marker of local intestinal inflammation 39,40 and has been associated with cancer progression and may influence tumor progression rather than tumor formation. 41. We observed participants with normal and elevated fecal calprotectin levels within each diagnostic group, and covariate-controlled analysis of the LCPM cohort revealed that 8 and 19 CRC-related markers were found in feces at the species and genus level, respectively. It was revealed that it was related to calprotectin in the medium, but not to calprotectin in the stool. diagnostic group. We repeated these observations in an independent cohort of apparently healthy individuals (FGFP). High levels of calprotectin in the stool are associated with inflammatory bowel disease19. However, when IBD patients were excluded from the analysis, the CRC diagnostic group remained non-significant, and the importance of Fusobacterium nucleatum among the other six species did not change after differential abundance analysis. In patients with colorectal cancer, elevated fecal calprotectin levels (>50 μg g g fecal 18,26) are directly related to the presence of tumor, as levels decrease after tumor resection 42. Here, fecal calprotectin is increased in CRC, consistent with previous associations between malignant transformation, local inflammation 43 and advanced tumor stages (T3 and T4) 42 . No difference in calprotectin levels was observed between CTL and ADE (mean 73.25 vs. 70.24 μgμgμg), indicating detectable levels of local inflammation despite the absence of lesions in the colon of the CTL group. It suggests that there is.
The potential of local inflammation on gut microbiota formation in the setting of malignant transformation is unclear, as most studies examining the association between gut microbiota and CRC13,14 have not controlled for local inflammation. effect, or its potential confounding effects, remain largely unknown. inflammation. To assess the potential clinical relevance of three species repeatedly found to be associated with CRC, including Escherichia coli, Fusobacterium nucleatun, and Parvimonas spp. , argue that strict control of covariates is essential in microbiome analysis. Although micros have been shown to be associated with local inflammation, they have unfortunately been uncontrolled in previous studies and may or may not be associated with cancer progression. Fusobacterium nucleatum is one of the species that has received more attention due to the large number of studies related to CRC44. In this study, Fusobacterium was enriched in colorectal cancer patients. However, this apparent association disappears once the analysis is controlled for covariates. Our study suggests that the association between Fusobacterium nucleatum and cancer may be driven by its association with inflammatory bowel disease.
Once calprotectin is controlled, there is no difference in the frequency of Fusobacterium nucleatum between diagnostic groups. These results suggest that the diagnostic utility of this marker will be reevaluated. At the same time, our results do not mean that Fusobacterium nucleatum is not associated with colorectal cancer. Rather, they suggest that the reasons for this relationship may not be as clear as initially thought. Thus, they are a cautionary tale about the importance of controlling for covariates as the field of microbiome advances. Given that inflammation is a risk factor for colorectal cancer but not its prerequisite,41 the possibility of using Fusobacterium nucleatum as a marker for colorectal cancer development may not identify cases of cancer progression that are not dependent on inflammation. Although not yet commercialized, there are already publications suggesting the use of microbial markers, including Fusobacterium nucleatum, for CRC screening,7,45 raising concerns in light of our results that uncontrolled variables may obscure the actual biological mechanisms. We hypothesize that putative CRC biomarkers, even those that have been replicated in multiple studies, may suffer from combined or confounding effects of covariates, which may lead to the use of non-quantitative signals. In addition, we present evidence that may lead to misleading conclusions about the actual diagnostic signal. This makes the path to potential clinical applications more difficult.
BMI is independently associated with changes in the gut microbiota, either in combination with inflammation or independently of inflammation 46 , which in turn is associated with increased risk of CRC 47 . However, microbial dysbiosis alone cannot explain the higher risk of colon cancer observed in obese populations, and the underlying processes linking obesity and colon cancer are more complex. This suggests that further investigation is required 48.
Of the four intestinal enterotypes described, the Bact2 enterotype has been defined as a dysbiotic microbial profile24,25. Enrichment of Bact2 has been observed in obesity25 and diseases such as PSC (primary sclerosing cholangitis) and IBD24, further supporting the possibility that this enterotype is associated with disease. Analysis of the LCPM cohort revealed an excess of Bact2 enterotypes in all diagnostic subgroups, regardless of BMI. The increase in Bact2 prevalence in the no-lesion group compared to the FGFP group is particularly striking. Patients in the CTL group have no detectable lesions but are at high risk for colorectal disease based on clinical evidence (e.g., blood loss in the stool, known risk of colonic involvement) that warrants colonoscopy. It's possible. This is also reflected in the Bact2 enterotype. Importantly, "healthy" biopsies included in CRC microbiome studies are often selected using colonoscopy with negative results as the main criterion, which poses a potential problem. . This is because other markers of colon health are not considered in determining eligibility for healthy people. Although the reasons for the appearance of Bact2 in the disease-free group vary, these results indicate that although such individuals are a useful category for biomarker discovery, they may harbor an unhealthy gut ecosystem from a microbial perspective. This suggests that there is a possibility that
There are various variables that have been identified as modifiers of the gut microbiota. Nevertheless, covariate control is far from standard and is not included in most relevant studies. As gut microbial taxa have been cited as potential biomarkers of malignant transformation, it is essential to investigate the influence of microbiota covariates as potential confounders or accelerators of the observed associations. is. Because these covariates alone can explain most of the variation in fecal microbiota independent of colorectal cancer status, our analysis does not negate previous associations, but rather supports clinical relevance. This emphasizes the need for covariate control analysis in microbiome studies aimed at establishing microbiota studies.
Of the numerous taxa previously associated with CRC, six remain significant after tightly controlling for covariates in this quantitative cohort. Although not ruling out other potential biomarkers, there have been no reports for Anaerococcus vaginalis, Dialister pneumosynthes, Parvimonas micra, Peptostreptococcus anaerobius, Prevotella intermedia, and Porphyromonas asaccharolytica. The association with CRC6,7 is strong enough to be method independent and warrants further study. Our data provide a strong basis for reconsidering potential associations between microbes and clinical phenotypes to ensure that they are not driven by uncontrolled covariates, and that these further tracing of the mechanisms underlying the association is warranted. Improved approaches to microbial biomarker discovery will undoubtedly impact the field of microbiome and facilitate the path to much-needed clinical applications.
Limit
Our aim is to identify taxa associated with malignant transformation of the colon. Although our cohort includes many participants without lesions, we cannot argue that these are healthy controls, as the incidence of dysbiosis appears to be increased in this group. I don't. Considering that all participants in this study had a medical need for colonoscopy, this implicitly increases the risk of colon cancer. Therefore, the present study cannot exclude the possibility of potential molecular or cellular alterations occurring in the polyp-free group that are not detectable by colonoscopy. Moreover, because this is a cross-sectional study, the term “cancer progression” is an extrapolation of what is observed in advanced cancer stages (operationalized here as diagnostic groups). Most studies did not provide sufficient metadata to allow for cohort comparisons, so we cannot exclude cohort peculiarities that may contribute to the observations. It is important to consider that certain taxa may not even be represented in the current databases, and that certain microbial species may require longer hypervariable regions or alternative sequencing approaches to achieve accurate species-level identification. Nevertheless, as we show using Fusobacterium as an example, in our cohort, the V4 region appears to be able to resolve species classification of biomarkers previously associated with CRC. Furthermore, the potential diagnostic value of colonic microbial profiles has been suggested to extend beyond bacteria, as fungal and viral species have been suggested as CRC biomarkers 49 . To provide comprehensive information for cancer detection and treatment, multidomain approaches to CRC biomarker discovery and prospective longitudinal studies to investigate the dynamics of cancer progression in more detail are needed. We recognize that it is necessary.
Method
Call for Participation The LCPM project was an observational cross-sectional study, and its procedures were approved by the UZL Medical Ethics Committee (ethics approval number S57084). Patients were recruited from 2017 to 2018 through research nurses using standardized procedures. Briefly, patients scheduled for lower gastrointestinal endoscopy or abdominal surgery for colorectal cancer resection at UZL were invited. After explaining the research project, if participants expressed consent, they signed an informed consent form and no remuneration was provided.
We have prepared a set of stool collection materials. Each patient completed a detailed questionnaire including, among others, information on sample collection date, stool consistency, diet, antibiotic use, clinical symptoms or diseases, and additionally an extensive medical and clinical questionnaire via the web-survey service KU Leuven. As a validation cohort, we included FGFP17, a population-scale microbiome surveillance effort, representing one of the largest and best-characterized fecal microbiome databases currently available. Its extensive metadata, including health and lifestyle, allowed the identification of 69 factors associated with microbiome variation (microbiome covariates). QMP transformation was performed in parallel using the same protocol for both the FGFP and LCPM cohorts. Classification of CRC status We invited patients referred for colonoscopy or colectomy to participate in the study.
Those who consented were asked to collect a stool sample at home, which was frozen using a sample kit provided by the research team. After completing a medically necessary procedure (colonoscopy or colectomy), study participants were divided into three diagnostic groups according to their clinical phenotype: (1) without signs of pathology; patients, (2) patients with polyps (nâ<â); 10, size 6–10 mm) (ADE) and (3) CRC patients. Patients whose clinical symptoms did not fall into any of these three groups were excluded from the study. Once participants were enrolled in the appropriate group, extensive metadata was collected from their medical records as indicated on the informed consent form. sample collection Stool samples from patients with UZL were collected as part of the LCPM project using aliquots of ready-made mats without buffers or preservatives (Supplementary Figure). 1). Samples were stored in the patient's home in her 20°C freezer and transported to the laboratory in ice packs. Upon arrival, samples were stored at 80 °C until further analysis in the Raes laboratory. Each stool sample was equipped with a temperature logger to ensure that a low and stable temperature was maintained during storage at home or transportation to the laboratory. Analysis of stool samples Measuring microbial load using flow cytometry We measured the microbial burden in stool samples of LCPM patients according to published methods23. For all other samples, cell counting was performed in triplicate. Briefly, a 0.2 g frozen (80°C) aliquot was dissolved in physiological solution to a total volume of 100 ml (8.5 g g l1 NaCl; VWR International). . The slurry was then diluted 1000 times. Samples were filtered using sterile syringe filters (5 μm pore size; Sartorius Stedim Biotech). Then, 1 ml of the resulting microbial cell suspension was stained with 1 ml of SYBR Green I (1:100 dilution in dimethyl sulfoxide, incubated in the dark at 37 °C for 15 min, 10,000 concentration, Thermo). Fluorescence events were monitored using FL1 533/530 nm and FL3 nm >670 nm photodetectors on a C6 Accuri flow cytometer (BD Biosciences) using a C6 Accuri flow cytometer (BD Biosciences). In addition, forward scattered light and side scattered light were collected.
Microbial fluorescence events in FL1/FL3 density plots were separated from background events using BD Accuri CFlow software (v.1.0.264.21). Supplementary Figure 2. A threshold of 2,000 was applied to the FL1 channel. Evoked fluorescence events were evaluated in forward and lateral density plots to exclude residual background events. Instrumentation and gate settings were kept the same for all samples as previously described24. Cell counts were converted to microbial load per gram of fecal material based on the exact weight of the analyzed aliquot. water content in stool We measured water content as a percentage of mass loss after freeze-drying 0.2 g of frozen aliquots (80 °C) of non-homogenized fecal material as previously described 24. Measurement of calprotectin in stool Fecal calprotectin concentrations were quantified using the fCAL ELISA kit (Buhlmann). For the patient and her FGFP participants, analysis of frozen fecal material (–80°C) was performed as previously described24. Phylogenetic profiling of the microbiome.
DNA extraction and sequence data preprocessing
The fecal microbiota profile of the FGFP cohort has been previously described. The same protocol was followed for fecal DNA extraction and microbiota profiling of the new cohort 17 . Bacterial profiling was performed as previously described 50. Briefly, nucleic acids were extracted from frozen stool aliquots using the MagAttract PowerMicrobiome DNA/RNA kit (Qiagen). We modified the manufacturer's protocol by adding a heating step of vortexing at 90 °C for 10 min and removing the step to remove DNA. For bacterial and archaeal characterization, we used 16S ribosomal RNA primers 515F (5â²-GTGYCAGCMGCCGCGGTAA-3â²) and 806R (5â²-GGACTACNVGGGTWTCTAAT-3â²) targeting the V4 region. These primers were modified to include a barcode sequence between each primer and an Illumina adapter sequence to create triplicate dual-barcoded libraries from extracted DNA (1:10 dilution). Deep sequencing was performed on a MiSeq platform (2 × 250 paired-end reads (PE), Illumina). All samples taken for sequencing and negative controls (polymerase chain reaction (PCR) and extraction controls) were randomized. After demultiplexing using sdm as part of the LotuS pipeline (version 1.60) without considering mismatches, Fastq sequences were further analyzed using the DADA2 pipeline (version 1.6). That is, the primer sequences and the first 10 nucleotides after the primer were removed. After merging paired sequences and removing chimeras, taxonomy was assigned using the formatted Silva set “SLV_nr99_v138.1”. Taxonomic assignments were made at domain, class, order, family, genus and species level using the “assignTaxonomy” function from the DADA2 R library, with a simple Bayesian classification method implemented by “silva_nr99_v138.1_wSpecies_train_set.fa” with a minimum bootstrap confidence of 50. gzâ training database (Extended Data Figure) Five). Deep sequencing his MiSeq of his DADA2 R library using the formatted Silva SSU database “silva_species_assignment_v138.1.fa.gz” to obtain species assignments of amplicon sequence variants (ASVs). executed on the platform. To avoid missing labels, we labeled all unassigned ASVs at each taxonomic level with the prefix “uc” along with the assigned taxonomic level (not the species level). Prior to analysis, sequences annotated with chloroplast class, mitochondrial family, or unknown archaeal and bacterial origins of eukaryotic origin were removed. philosique (v. The 1.36.0)53 and MicroViz (v. 0.11.0)54 libraries were used for data curation and figure generation. RMP For relative microbiome matrices, ASV counts were converted to relative abundances. In other words, we divided the number of ASVs by the total number of ASVs per sample. ASVs were aggregated to the species level using phyloseq (v.). 1.36.0)53 "tax_glom" function. RMP (CLR) ASVs were aggregated to the species level and abundance matrices were centrally logarithmically (CLR) transformed using “codaSeq.clr” in CoDaSeq (v. 0.99.6), determined for each individual taxon for imputation zero. Minimum proportional abundance was used. Workflow evaluation We performed workflow evaluation using (1) ZymoBIOMICS Gut, a commercial mock community, and (2) two Fusobacterium species, Fusobacterium hwasookii (THCT14E2) and Fusobacterium nucleatum (DSM 20482T). The evaluation followed our standard methodology and included amplification, sequencing, and analysis of extracted DNA. The purpose of this evaluation was to assess the performance of the overall methodology, as shown in Figure 6 in the extended data. Quality control evaluation of amplicon sequence data (16S rRNA) using RMP Briefly, all samples were sequenced in six MiSeq runs (Extended Data Figure 7a). For each run, a series of internal controls were used to detect: 1) intra-run and between-run technical variations; 1) contamination events during DNA extraction; 2) contamination events during amplification and sequencing steps; and 3) crosstalk between contaminated equipment and barcodes due to sequence carryover. Biological materials (stool samples), positive controls (DNA and RS from previously profiled stool samples: non-human enterobacterial strain 'Runella Srithyformis'), negative controls (negative controls extracted (NCE) and negative controls) We amplified all samples containing A unique barcode combination was used to provide control during the three PCR (NCP) runs, while omitting multiple barcode combinations to control cross-contamination due to primer synthesis. To detect barcode crosstalk, we replicated Runella Srithyformis in each sequencing library (Extended Data, Figure 7b). Since this genus is not detected in human intestinal samples, we expected that Lunella thuriformis levels would not be detected in the stool samples analyzed (extended data). fig. 7c). Finally, we incorporated NCE along the entire process from extraction to bioinformatics analysis. We used NCP and NCE (Extended Data Figure 7d and Supplementary Table 12) for contamination amplification and sequencing, and for permeation contamination events we used a set of different barcode combinations in consecutive MiSeq runs56. QMP A QMP matrix was created as previously described23. Briefly, uniform We reduced the sample to the sampling depth. We calculated 16S rRNA genome copy number (GC) using RasperGade16S (v. 0.0.1)57, a novel tool that predicts 16S rRNA GC using a heterogeneous pulse evolution model. It not only predicts GC but also provides a confidence estimation of the prediction57. A minimum number of reads less than 150 was used for QMP analysis. The final size of the QMP matrix, which converted diluted ASV abundance to cells per gram, was 589 samples for the study cohort and 1,045 samples for the FGFP validation cohort. . We use the Phyloseq (v. 1.36.0)53 function “tax_glom” to aggregate the ASV-level QMP matrix down to the species level. The resulting species QMP matrix was used for the main analysis. statistical analysis All statistical analyzes were performed using R (version 4.2.1, RStudio v.2022.12.0â+â353, 86_64-apple-darwin17.0 (64 bit)) and package Phyloseq (v. 1.36.0). did. 53, vegan (v. 2.6.2)58, coin(v. 1.4.2)59, effect size (v. 0.8.3), vcd(1.4.11)60, DirichletMultinomial(v. 1.34.0)61, pairwiseAdonis (v. 0.4. 2) 1) and the microbiome (V. 1.14.0)62. Non-parametric statistical tests were used for robust comparisons between unbalanced groups. For multiple testing, all P values were corrected using the Benjamini-Hochberg method (reported as adjusted P) according to the list of features (nâ>â) 1) (taxon and metadata or metadata and (e.g. metadata association). Perform some pairwise group comparisons (n≠2) (e.g. his KW test with the PhD test).
Characteristics and visualization of fecal microbiota
We visualized interindividual variation in the microbiota by PCoA using his BCD on a species QMP matrix. All other microbiota-derived traits were calculated based on QMP. We determined the contribution of metadata variables to microbiota community variation (effect size) for each of the 94 variables by dbRDA on species-level BCD using the cap-scale function of the vegan package58. We visualized the absolute abundance of species as log10 (abundance +1). This also applies to relative frequencies. Relationship between microbiota and physiological traits
Taxa that were not classified to the species level or that were present in less than 5% of samples per diagnostic group were excluded from the analysis (Supplementary Table 6). Spearman correlation was used for rank correlations between continuous variables such as species abundance, calprotectin levels, and water content, supplemented with Kendall's tau correlation. The Mann-Whitney U test was used to test for differences in median values of continuous variables between two different groups. For more than two groups, for example, for differential abundance analysis of QMP and RMP taxa compared to diagnostic groups, we used KW and PhD tests. ANOVA tests were performed for differential abundance analysis between diagnostic groups and analysis of bacterial species abundance from CLR-transformed data. Pairwise CS tests were used to assess statistical differences in the proportions of categorical variables (enterotypes) between patient groups. We tested whether there was a contribution of the decoded microbiota to the diagnostic group variables using generalized linear model nested model comparisons (ANOVA) as follows.
$$\begin{array}{l}[{\rm{null}}\,{\rm{model}}]\,{\rm{glm}}0={\rm{rank}}({\rm {frequency}})+{\rm{rank}}({\rm{calprotectin}})\\\qquad\qquad\qquad\quad\quad+{\rm{rank}}({\rm{moisture} }) +{\rm{Rank}}({\rm{BMI}})\end{Array}$$
[Alternative model]glm1â=ârank(abundance)â+ânk(calprotectin)â+ârank(moisture)â+ârank(BMI)â+âDiagnosis, diagnostic groups include patients without signs of her CTL, patients with polyps, and A patient with CRC. , were recoded as 1, 2, and 3, respectively. We treated this variable as a continuous variable and translated the directional increase in disease progression from a healthy state to a diseased state in the colonic mucosa. Nested model comparisons used taxon abundance (quantitative or relative) as explanatory variables, diagnostic group variables as response variables, and BMI, fecal calprotectin, and humidity as covariates. Additionally, we used rank transformation modeling to perform nonparametric tests on non-normally distributed data, including: B. species richness. Previous reports on microbial CRC markers
We used the keywords “CRC AND microbiome AND stool AND human AND biomarker” to create a list of published CRC markers that define taxa to test against covariates in the dataset. A PubMed search was performed. We found 10 studies that met our inclusion criteria. (1) sample size of at least 60, and (2) CRC biomarkers described at the species level with statistical significance in the text of the publication. We included this list of published biomarkers in a correlation analysis between taxa and three key covariates (fecal calprotectin, BMI, and humidity) within the LCPM cohort. A similar procedure was used at the genus level, and 15 studies found in the PubMed search were included.
Identification of microbial CRC markers
We conducted differential frequency analysis on nine different CRC shotgun datasets as part of the curated MetagenomicData33. This analysis used the MetaPhlAn 3.0 profile to compare the results and control for potential differences from the classification tools and statistical tools used in the results of each independent study method. The results of the meta-analysis are shown in Extended Data Figure 8 and Supplementary Table 13.
Enterotyping and visualization
We calculated the observed genus richness by enterotyping using a genus matrix (aggregated and downsized to 10,000 reads) as reported in a previous study. For enterotyping (or community typing) based on the DMM approach, we used R as previously described 61 . We performed genus-level enterotyping of a combined RMP matrix containing LCPM samples compiled from 1,045 samples derived from FGFP17. The optimal number of Dirichlet components based on the Bayesian information criterion was four. As previously mentioned, the four clusters were named “Bact1,” “Bact2,” “Prev,” and “Rum.”23
Report summary
For more information on the study design, please see the Nature Portfolio Reporting Summary linked in this article.



إرسال تعليق